sparklyr is a great opportunity for R users to leverage the distributed computation power of Apache Spark without a lot of additional learning. sparklyr acts as the backend of dplyr so that R users can write almost the same code for both local and distributed calculation over Spark SQL.
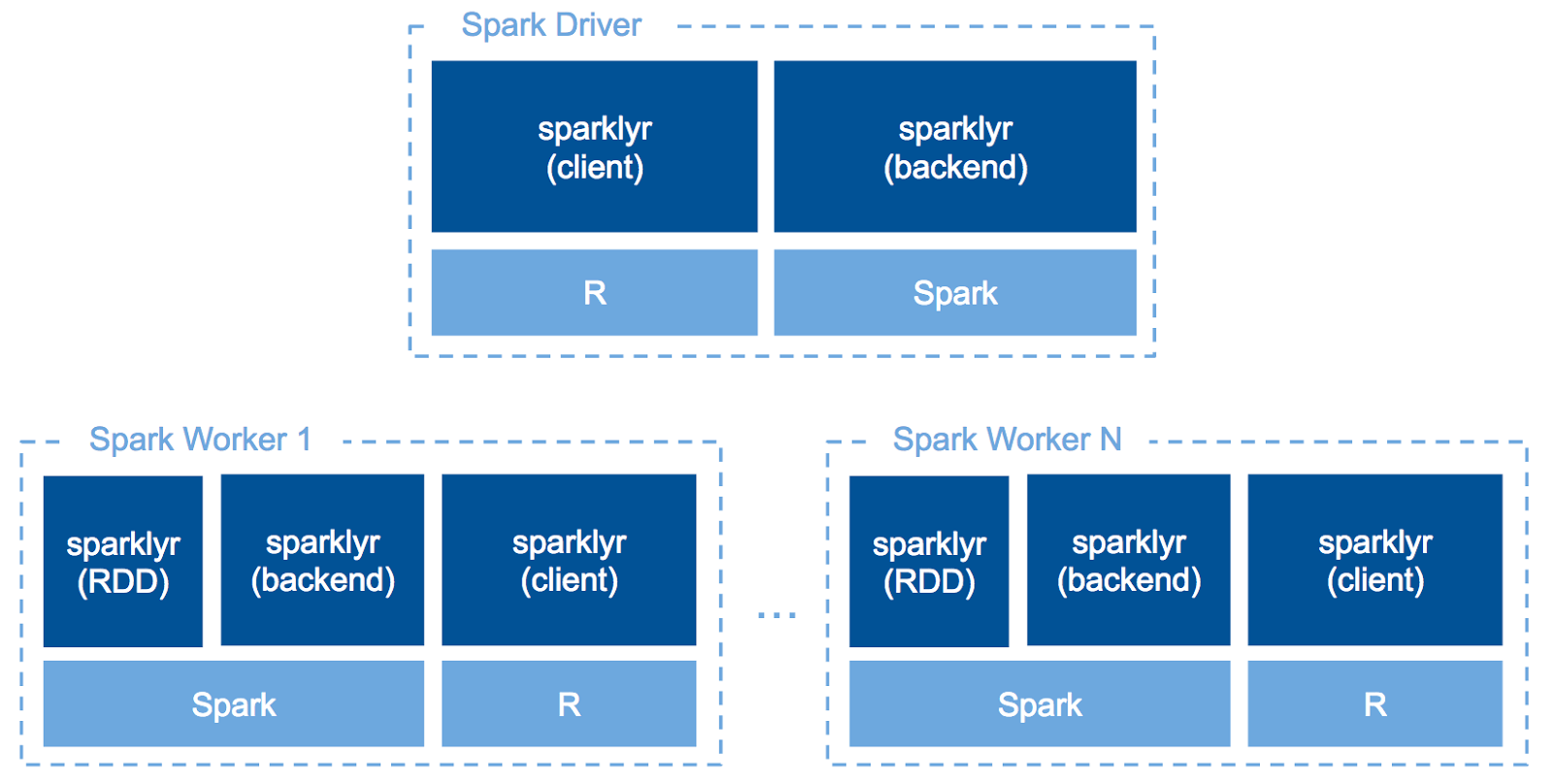
Since sparklyr v0.6, we can run R code across our Spark cluster with spark_apply(). In other words, we can write UDF in R. This makes you to use your favorite R packages for data on a Spark cluster to use specific statistical analysis methods only implemented in R or some advanced analysis like NLP, etc.
Since the current implementation of spark_apply() requires R on worker nodes, in this post, we will introduce how to run spark_apply() on CDH clusters. We will explain two ways to achieve executing it: 1) Using Parcel 2) Using a conda environment.
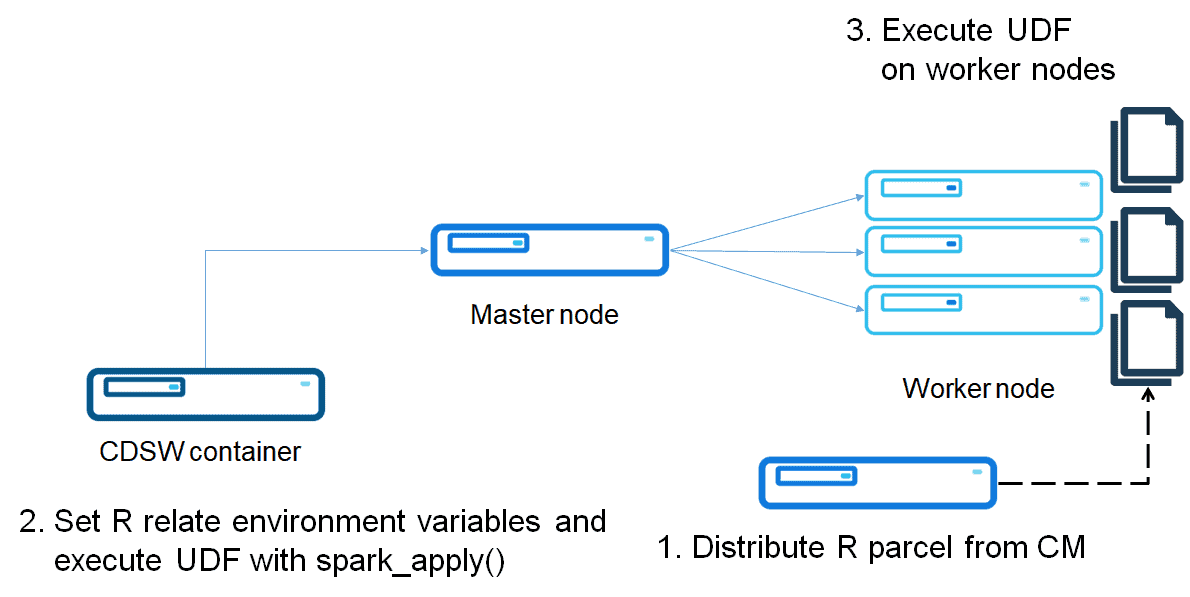
Option 1: Install R environment with Parcel
Build and distribute an R Parcel
Parcel is a binary distribution format and Cloudera Manager supports it to distribute CDH, Spark 2, Kafka and other services running on the cluster. The purpose of a Parcel is similar to that of a .deb or .rpm package. It enables you to install a particular program on a CDH cluster easily via the Cloudera Manager UI. This option assumes the cluster is to be deployed by Parcel-based distribution.
You can download pre-built parcels from Bintray, which includes basic components powered by the r-essentials recipe of conda. Note that you can’t set Bintray as a remote Parcel repository directly, so you should upload those parcels on to your HTTP server.
If you need to use other R packages, use the parcel build tool for R. You can build your parcels with Docker by modifying a Dockerfile.
https://github.com/chezou/cloudera-parcel
Put the parcels on an HTTP server or a given S3 bucket. Then you can add the parcel repository URL on Cloudera Manager. See this document for more detail.
Run R code on Spark worker nodes
After distributing the R parcel, you can run R code on worker nodes.
Note: To set environment variables, currently, we need to use the upstream version of sparklyr. The latest release version v0.6.1 doesn’t have this function.
Here is the example repository for distributing R code.
https://github.com/chezou/sparklyr-distribute
devtools::install_github("rstudio/sparklyr")
library(sparklyr)
config <- spark_config()
config[["spark.r.command"]] <- "/opt/cloudera/parcels/CONDAR/lib/conda-R/bin/Rscript"
config$sparklyr.apply.env.R_HOME <- "/opt/cloudera/parcels/CONDAR/lib/conda-R/lib/R"
config$sparklyr.apply.env.RHOME <- "/opt/cloudera/parcels/CONDAR/lib/conda-R"
config$sparklyr.apply.env.R_SHARE_DIR <- "/opt/cloudera/parcels/CONDAR/lib/conda-R/lib/R/share"
config$sparklyr.apply.env.R_INCLUDE_DIR <- "/opt/cloudera/parcels/CONDAR/lib/conda-R/lib/R/include"
sc <- spark_connect(master = "yarn-client", config = config)
sdf_len(sc, 5, repartition = 1) %>%
spark_apply(function(e) I(e))r
## # Source: table<sparklyr_tmp_1cc757d61b8> [?? x 1]
## # Database: spark_connection
## id
## <dbl>
## 1 1
## 2 2
## 3 3
## 4 4
## 5 5
If you want to use R packages in a distributed function, sparklyr packs those packages under local .libPaths() and distributes them to each worker node using SparkContext.addFile() function. If you are using packages within spark_apply() that depend on native code, you can distribute them using Conda as well as described in the next section.
See more detail in the official document: https://spark.rstudio.com/guides/distributed-r/#distributing-packages
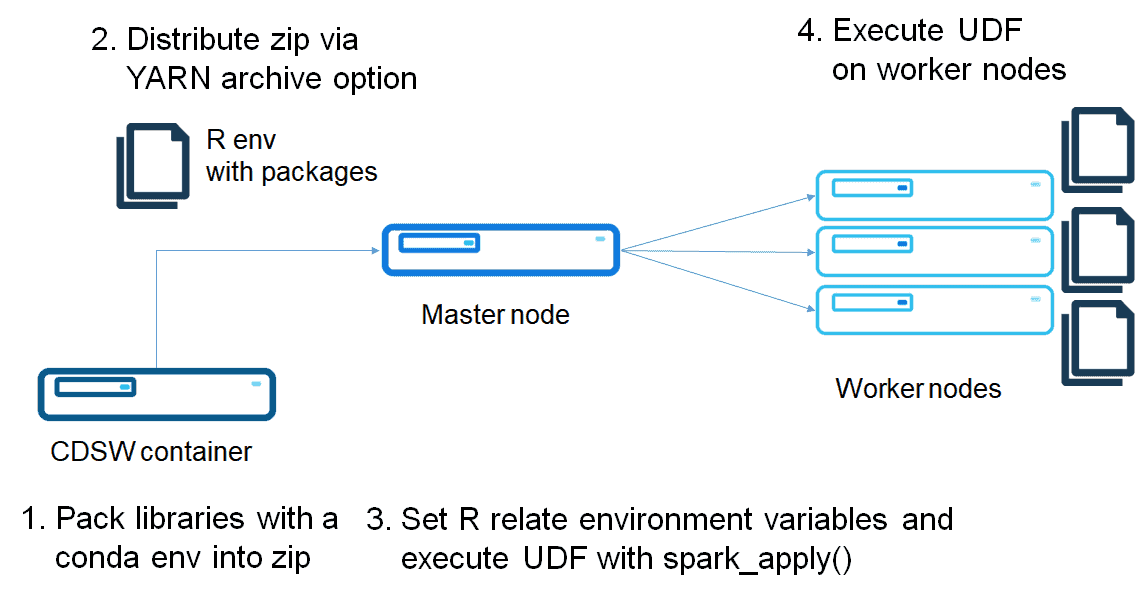
Option 2: Use conda environment
Build a conda environment with R
As I mentioned in the previous post, a conda virtual environment can package an R environment.
Create a conda environment of R, and zip it as follows:
$ conda create -p ~/r_env --copy -y -q r-essentials -c r
# [Option] If you need additional package you can install as follows:
# $ source activate r_env
# $ Rscript -e 'install.packages(c("awesome-package"), lib = /home/cdsw/r_env/lib/R/library, dependencies = TRUE, repos="https://cran.r-project.org")'
# $ source deactivate
$ sed -i "s,/home/cdsw,./r_env.zip,g" r_env/bin/R
$ zip -r r_env.zip r_env
The differences versus using Parcel are the environment variable setting and setting r_env.zip as an environment variable. While this is flexible method, this method requires distributing a zip file each time you create a Spark connection. Full code is in the following repository: https://github.com/chezou/sparklyr-distribute/blob/master/dist_sparklyr_conda.r
config <- spark_config() config[["spark.r.command"]] <- "./r_env.zip/r_env/bin/Rscript" config[["spark.yarn.dist.archives"]] <- "r_env.zip" config$sparklyr.apply.env.R_HOME <- "./r_env.zip/r_env/lib/R" config$sparklyr.apply.env.RHOME <- "./r_env.zip/r_env" config$sparklyr.apply.env.R_SHARE_DIR <- "./r_env.zip/r_env/lib/R/share" config$sparklyr.apply.env.R_INCLUDE_DIR <- "./r_env.zip/r_env/lib/R/include"
Then, you can run R code on Spark worker nodes.
Complex example: Text analysis with spacyr
NOTE: Please notice that using R packages with native code within spark_apply() is currently unsupported in this release.
In this example, we use spacyr package, which is R binding of spaCy; new Python NLP library. We extract named-entity, such as person, place, organization, etc. from Jane Austen’s books. Full code of this example is here.
https://github.com/chezou/spacyr-sparklyr
Prepare conda environment for spacyr
Since spacyr requires Python runtime, you should install the Anaconda parcel on the cluster before running this example.
You should create conda environment with this script.
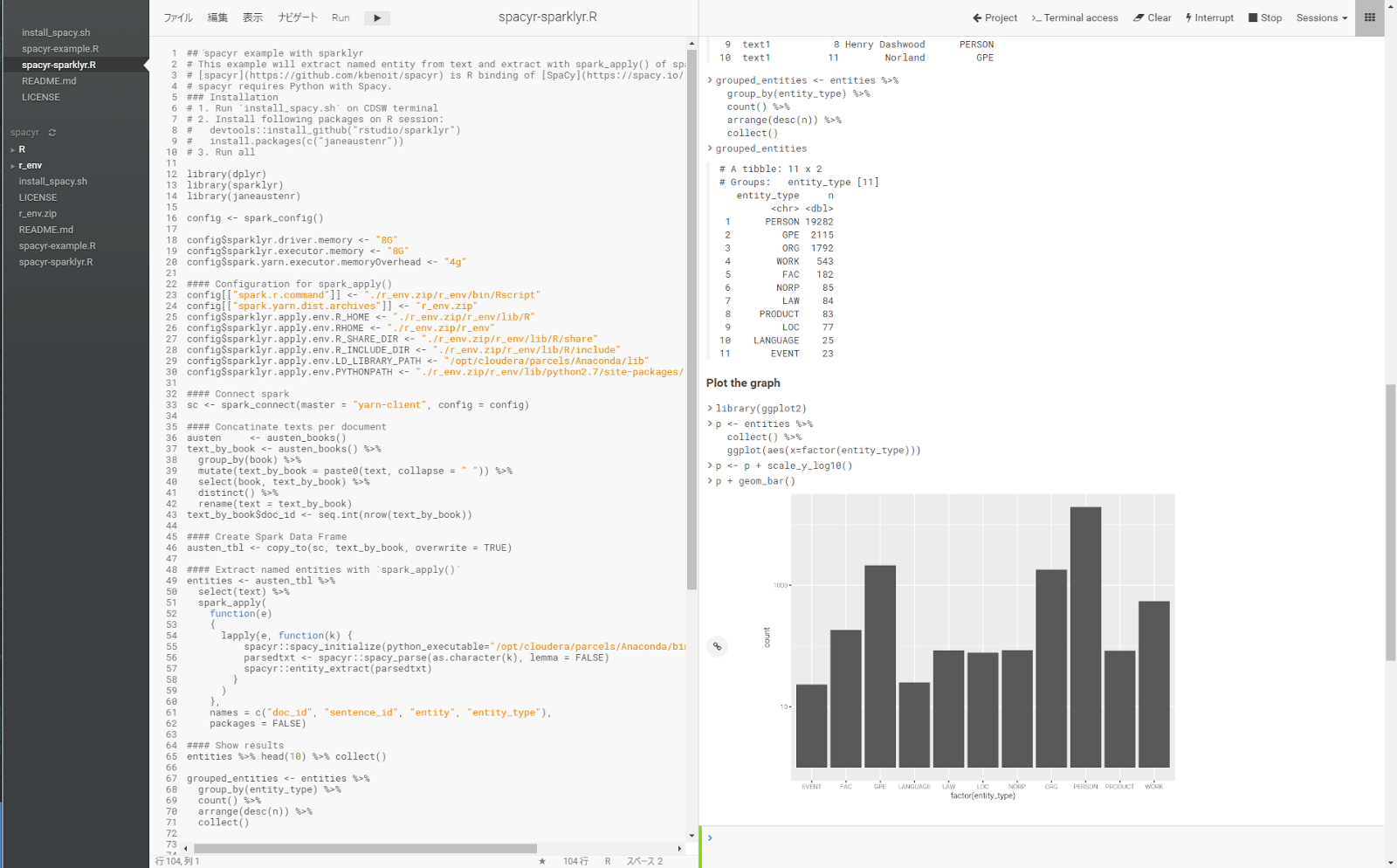
Extract named entities with spark_apply()
With Spark DataFrame having text column, you can We can extract named-entity with following UDF:
entities <- austen_tbl %>%
select(text) %>%
spark_apply(
function(e)
{
lapply(e, function(k) {
spacyr::spacy_initialize(python_executable="/opt/cloudera/parcels/Anaconda/bin/python")
parsedtxt <- spacyr::spacy_parse(as.character(k), lemma = FALSE)
spacyr::entity_extract(parsedtxt)
}
)
},
names = c("doc_id", "sentence_id", "entity", "entity_type"),
packages = FALSE)
entities %>% head(10) %>% collect()
## # A tibble: 10 x 4
## doc_id sentence_id entity entity_type
## <chr> <int> <chr> <chr>
## 1 text1 1 Jane Austen PERSON
## 2 text1 1 Dashwood ORG
## 3 text1 1 Sussex GPE
## 4 text1 2 Norland Park GPE
## 5 text1 4 Henry Dashwood PERSON
## 6 text1 4 Norland GPE
## 7 text1 5 Gentleman NORP
## 8 text1 7 Henry Dashwood PERSON
## 9 text1 8 Henry Dashwood PERSON
## 10 text1 11 Norland GPE
Let’s plot the number of entity types.
library(ggplot2) p <- entities %>% collect() %>% ggplot(aes(x=factor(entity_type))) p <- p + scale_y_log10() p + geom_bar()
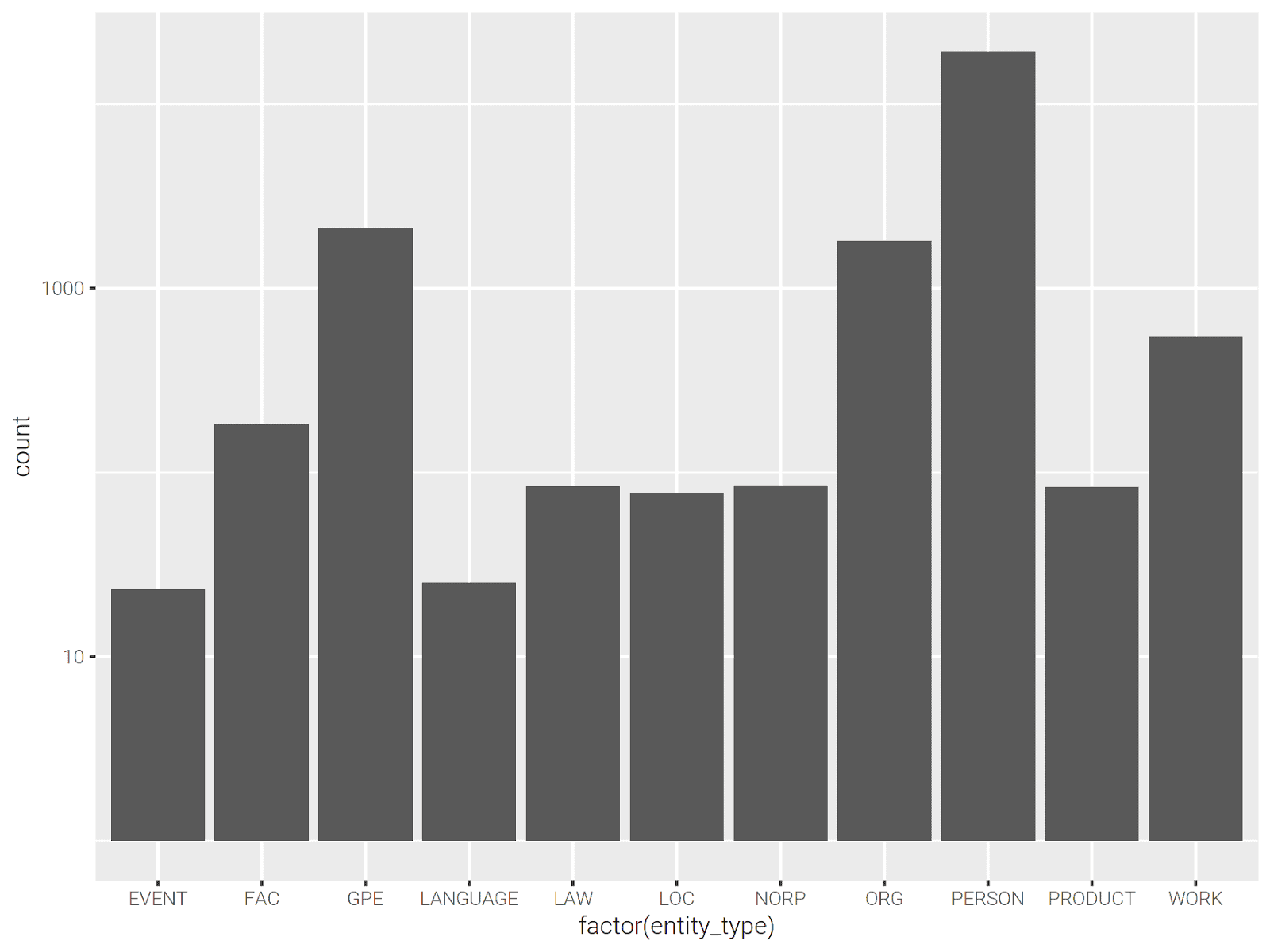
We can see top 10 frequent people for each book as follows:
persons <- entities %>% filter(entity_type == "PERSON") %>% group_by(doc_id, entity) %>% select(doc_id, entity) %>% count() %>% arrange(doc_id, desc(n)) persons %>% filter(doc_id == "text1") %>% head(10) %>% collect() ## # A tibble: 10 x 3 ## # Groups: doc_id, entity [10] ## doc_id entity n ## <chr> <chr> <dbl> ## 1 text1 Elinor 662 ## 2 text1 Marianne 538 ## 3 text1 Edward 235 ## 4 text1 Jennings 233 ## 5 text1 Willoughby 200 ## 6 text1 Lucy 172 ## 7 text1 Dashwood 159 ## 8 text1 Ferrars 104 ## 9 text1 Lady Middleton 80 ## 10 text1 Palmer 74
Which option should I use?
Basically, I would recommend using Option 1, because you don’t need to distribute R environment each time, and building your own Parcel including all packages reduces a lot of time to struggling with package building. Currently, RStudio requires a Homogeneous Cluster, but you could work around the OS difference with packages = FALSE option.
On the other hand, option 2 is flexible to install packages lazily, but it could be complicated. If you want to use only pure R packages, this method would be straightforward, while if you need native extension packages such as rJava, it could be tough to prepare the environment.
Conclusion
We introduced how to run and distribute R code on Spark worker nodes with sparklyr. Since spark_apply() requires to be installed R on worker nodes, we explained two easy ways to run spark_apply() on a CDH cluster and CDSW. You can choose a preferred option with your purpose. If you need stability, you can use Option 1: parcel method. If you want to get flexibility, you can use Option 2: conda environment method.
You can leverage not only dplyr like execution, but also distribute local R codes on Spark cluster. It makes you take full advantage of your R skill on distributed computing.
Aki Ariga is a Field Data Scientist at Cloudera and sparklyr contributor